Paper: Analysis of BRAF Mutation in a Cohort of Patients with Glioblastoma Using Tissue Microarrays
High School Science Research Paper-- Weill Cornell Medicine
Introduction
Glioblastoma multiforme is the most malignant astrocytic neoplasm that arises in the human central nervous system. These WHO grade IV tumors can affect people of all ages, but are typically seen in older patients. The average age at diagnosis is 55 years (Ellinson, 2013). The incidence of glioblastoma is 2 to 3 cases per 100,000 adults annually. They make up 17% of all brain tumors, including metastases, but 52% of all primary brain tumors (AANS, 2015). Glioblastomas are aggressive and the prognosis is poor. Average survival following the diagnosis of a glioblastoma multiforme is just 14 months (Ellinson, 2013). Current treatment methods are ineffective and have low success rates. Patients have very few options and unfortunately most succumb to their disease in a short period of time.
The current standard of care for patients diagnosed with a glioblastoma is a surgical resection, followed by six weeks of radiation and temozolomide chemotherapy (Prados et al., 2015). Surgery is an option for many patients, but it is in no way a cure. Extensive surgical procedures may be performed to eliminate all visible traces of the malignancy, but due to the infiltrative nature of the disease, residual tumor is almost always left behind (Prados et al., 2015). Through histological analysis it is evident that tumor cells permeate through the healthy tissue surrounding the tumor and cannot be completely resected without the risk of neurological deficits. The remaining tumor cells continue to proliferate and cause a rapid recurrence of the disease in many patients. Recurrent tumor is typically discovered six to nine months after the initial diagnosis (Prados et al., 2015). Even the most promising clinical trials do not prolong the survival of patients. Recent research has been moving toward precision medicine.
Precision medicine involves identifying specific mutations in an individual’s cancer and using targeted therapies that match these alterations. There are a variety of gene mutations that are present in astrocytic tumors such as glioblastoma. Glioblastomas appear in many forms, and the histology and the genetic alterations can vary between subtypes or variants. One mutation present in a relatively small number of glioblastomas, but is of recent interest is the BRAF V600E point mutation. BRAF is part of the receptor tyrosine kinase RAS-RAF-MEK-ERK signaling pathway, which regulates cell processes such as proliferation (Nicolaides et al., 2011). Mutations of the BRAF gene activate the pathway and stimulate cell growth (Brandner et al., 2015). The BRAF V600E point mutation occurs at codon 600, where the change from T to A results in the exchange of amino acid valine for glutamine (Schinder et al., 2011). This specific BRAF point mutation has been discovered to be present in 7-15 % of all human cancers (Brandner et al., 2015). The point mutation is most commonly seen in melanoma but recently it has been discovered in a variety of other types of cancers, including gliomas (Schinder et al., 2011).
Previous studies have determined the incidence of the BRAF V600E mutation in a variety of central nervous system tumors. The BRAF mutation appears in about 5% of adult high grade gliomas, a category which encompasses glioblastomas (Schinder et al., 2011). A study in 2013 by Gutmann et al., found that 7.7% of the adult glioblastomas in their cohort of patients were BRAF V600E positive. Although the incidence rate of the BRAF V600E mutation is low in glioblastomas, the mutation is more common in specific groups of patients. In nervous system tumors, the incidence of the BRAF mutation is associated with younger patient age (Schinder et al., 2011). A study found that the BRAF V600E mutation is found in roughly 10% of the high grade pediatric malignant astrocytoma in two patient cohorts (Nicolaides et al., 2011). In addition to being more frequent in pediatric patients, the BRAF V600E point mutation is more common in a specific variant of glioblastoma. It has been discovered that the BRAF mutation appears in up to 50% of epithelioid glioblastoma (Gutmann, et al., 2013). This variant of glioblastoma is characterized by sheets of small cells that very closely resemble epithelial cells with rounded cell borders and clearly defined nucleoli (DeMasters et al., 2013). No histological features have been found to differentiate epithelioid glioblastomas that possess the mutation and those that do not (DeMasters et al., 2015).
Although the incidence rate of the BRAF V600E mutation in glioblastomas is relatively low, it is of interest because there is a targeted treatment available for this mutation. PLX4032, or vemurafenib, is a small molecule inhibitor that restricts the pathway containing BRAF (Nicolaides et al., 2011). Clinical trials began in 2008, and in 2011 vemurafenib was approved by the food and drug administration, specifically for the treatment of melanoma (Niezgoda et al., 2015). Trials of vemurafenib in advanced melanoma have been successful, with high response rates and few patients even showing a complete response (Niezgoda et al., 2015).
More recently, vemurafenib has been used to treat patients with other types of cancer that contain the BRAF V600E mutation, in particular, glioblastoma. In BRAF V600E mutated cell lines treated with PLX4032 and PLX4720, a similar inhibitor, cell growth rate was significantly decreased (Nicolaides et al., 2011). A pediatric patient with a BRAF mutated epithelioid variant glioblastoma had a recurrence after he received the standard of care. Surgery was determined to be a poor option for the treatment of the recurrent tumor. The patient was started on vemurafenib treatment and in the following months his radiological results showed first a partial then a complete response (Robinson et al., 2014). Another patient had a similar recurrence of their disease, and underwent a second surgical resection followed by vemurafenib inhibitor treatment. 23 months later, no signs of recurrent tumor have been seen (DeMasters et al., 2015).
The presence of the BRAF V600E mutation in glioblastoma has been a recent discovery. Even after the mutation had been discovered, not every glioblastoma patient has been screened. The mutation appears in relatively few glioblastoma. However, identifying the mutation, even in a small number of patients is very significant. Diagnosing the BRAF mutation provides a new treatment, that has been successful in many cases, for patients that have very few options.
Purpose
The purpose of my project was to identify the incidence of the BRAF V600E mutation in a cohort of glioblastoma using a tissue microarrays and BRAF V600E immunohistochemistry. Tissue microarrays are a cost and time effective method of screening for mutations. Once this method has been used to identify a mutation, sequencing can be used to confirm the positive results. The discovery of BRAF V600E mutations would provide a new treatment option for those patients who are currently alive with the mutation, in the likely event that they have a recurrence. Finding patients that have the mutation would validate screening all glioblastoma cases for the BRAF mutation by immunohistochemistry.
Materials and Methods
Retrieving Case Information
I first went through 208 high grade glioma cases pulled from the archives of the institution where I worked. The cases were selected from the years 2010 to 2014. Each case contained the specimens that had been used to diagnose the patients. This included blocks of formalin-fixed paraffin embedded tissue, and corresponding cuts of the tissue stained with hematoxylin and eosin. I recorded clinical information for each of the cases in a spreadsheet. The information included the case code, age, gender, location of tumor, diagnosis, if it was recurrent, and the immunohistochemistry results of Ki-67, IDH1, EGFR, and P53.
Selecting Optimal Cases for the Tissue Microarray
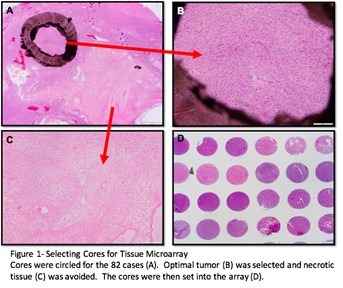
The next step was to create tissue microarrays of high grade glioma cases. A tissue microarray allows for three small tissue samples from up to 40 cases to be placed on one slide. This method makes staining more time and cost efficient. Tissue microarrays also enable researchers to easily compare cases and screen many cases at once.
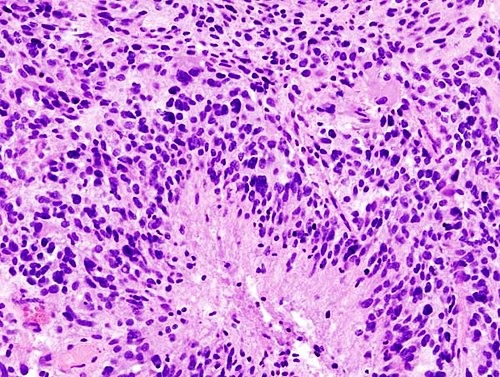
I first eliminated the cases that were not suitable for the array. Using a microscope and the hematoxylin and eosin stained slides from each case, I identified and separated out the cases that had little tissue left, minimal tumor, and necrotic or cauterized tissue, leaving only the cases with optimal tumor for coring. For each of the cases with sufficient tumor, I then examined and selected three 1mm areas that were best representative of the tumor. I identified the optimal tumor by scanning for the key characteristics of glioblastomas, which include pseudopalisading necrosis, microvascular proliferation, hypercellularity, atypical cells, and mitotic figures. When circling tumor to be cored, I also ensured each block of fixed tissue that corresponded to the slide I chose had sufficient tumor remaining to be cored for a tissue microarray. I added the tissue microarray number, block number, number of cores, and a any comments to the spreadsheet of the cases I selected. The circled tissue was microdissected and made into tissue microarrays by the Translational Research Lab technicians.
Analyzing Hematoxylin and Eosin Stained Tissue Microarrays
When I received the cut sides of the tissue microarrays that I had created, I selected the cuts that were most intact. I had one slide of each of the arrays stained with hematoxylin and eosin, so that I would be able to examine the histology and condition of the cores. I made a new spreadsheet with the core numbers of the array and their corresponding cases. For each core I examined with the microscope, I rated the cellularity, necrosis, presence of inflammatory cells, presence of necrosis, presence of neurons, microvascular proliferation on a scale from zero to three. I then used this information to give the overall condition of the core a numerical value. Zero indicated a missing core, one a poor core, two a good core, and 3 an excellent core.
Analyzing BRAF V600E Immunohistochemistry Stained Tissue Microarrays
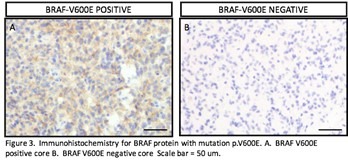
After examining the histology of the cores, I started to analyze them for the BRAF V600E mutation. I had them stained for the mutation by immunohistochemistry. I identified the cores that were positive for the mutation. I included all of the cores that were positive throughout the core or had small areas of BRAF V600E positive cells. I matched the cores to their corresponding cases and set these aside for further review.
Analyzing BRAF V600E Immunohistochemistry Stained Slides and Sequencing
After identifying the cases that were BRAF V600E positive by immunohistochemistry of the tissue microarray, I set out to confirm these results. First I had a full slide of tumor from each of the cases that were positive for the mutation by immunohistochemistry stained as well. This would confirm the positive result for the cases that had smaller areas of positivity.
In addition, I had the slides sequenced using next generation sequencing for the BRAF V600E mutation. Sequencing verifies the presence of the BRAF mutation and also provides additional data on other common genetic mutations that may be present in these tumors. Immunohistochemistry, while it is a time and cost efficient option, sometimes yields false positives as stains are developed and revised. The most efficient way to identify the mutation is to preliminary screen for the mutation then ensure the results with sequencing.
Additional Clinical Information on Possible BRAF Patients
Last, I obtained additional clinical information on the patients that were positive for the BRAF mutation by immunohistochemistry. This included their survival status and the other information on the presentation of their disease.
Results
Characteristics of the Cohort of High Grade Glioma Patients
I created two tissue microarrays with the cases that I selected. The first contains three cores of tissue from 42 patients and the second contains three cores of tissue from 40 patients, for a total of 82 patients. They were from the years 2010 to 2014 and ranged from ages 9 to 86. The average age was 57.5 years at the time of surgical resection and diagnosis of glioblastoma. There were 37 female patients and 45 male patients in the cohort. Nine of the cases were diagnosed as anaplastic astrocytoma (Grade III high grade astrocytic neoplasm), two were just diagnosed as high grade glioma, 70 of the cases were glioblastoma, and one was characteristic of both glioblastoma and PNET. 19 of the cases were recurrent tumor.
Quality of Tissue Microarray Cores
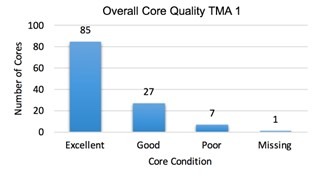
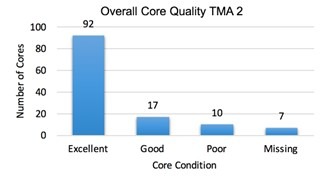
I assessed the quality of the tissue microarrays that I compiled by examining the cores using a hematoxylin and eosin stained slide. In the first TMA, there were 92 excellent cores, 17 good, 10 poor, and 7 missing. 86.5% of the cores were of good or excellent quality. The second TMA had 85 excellent, 27 good cores, 10 poor, and 1 missing core. 91% of the cores in the second TMA were of good or excellent quality. The tissue microarrays had very few missing cores and most of the cores were optimal. The tissue microarrays that were created are a viable way to screen for the BRAF mutation.
BRAF V600E Positive Cases by Immunohistochemistry
I assessed the BRAF immunohistochemistry stained tissue microarray, and were various degrees of positivity seen. Some cores had only a blush and others were positive throughout. The cases that had any sign of positivity are considered BRAF V600E positive by immunohistochemistry. There were a total of four cases that I found to be positive in the tissue microarrays. These cases were 19714, 19441, 29859, and 6456. The entire slides of these cases stained for BRAF by immunohistochemistry also showed varying degrees of positivity.
BRAF V600E Mutation Sequencing Results
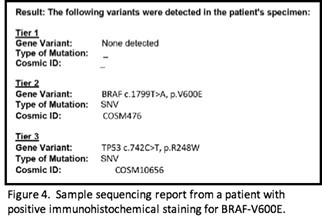
I had the four cases that were BRAF V600E positive by immunohistochemistry sequenced by next generation sequencing. The sequencing results include a gene panel of 50 common genetic mutations. Sequencing results revealed that two of the cases that yielded positive immunohistochemistry results actually had the mutation. These two cases were 19714 and 6456.
Clinical Information of BRAF Positive Cases
Patient 1
This patient has the BRAF V600E point mutation, screened by immunohistochemistry and verified with next generation sequencing. The patient was a 41 year old female at the time of diagnosis. The tumor was resected in 2012 and was diagnosed as a WHO Grade IV Glioblastoma Multiforme, located in the left frontal parietal lobe. The tumor was hypercellular, had mitotic activity, pseudopalisading necrosis, and microvascular proliferation, all characteristic of high grade glioma. The tumor diagnosis by the GFAP testing, which confirmed that the tumor cells were glial. The patient turned up negative for the IDH1 R132H mutation. The ki67 staining revealed a proliferation rate of up to 30% of the cells. The tumor was also positive for EGFR. P53 staining was variable in the tumor, but was positive in roughly 50% of the nuclei of the tumor cells.
This year, three years after the original diagnosis, the patient is alive. Following the surgery, the patient was treated with radiation and Temodar. An area of enhancement has appeared since the surgery in the left posterior ventricle. The small size of the lesion and the fact that it did not change in the following months has suggested that it was an effect of the radiation treatment and not a recurrence of the tumor. The patient is currently doing well and is not experiencing any symptoms.
Patient 2
This the second patient that is positive for the BRAF V600E point mutation, by immunohistochemistry and sequencing. The patient was a 78 year old female at the time of the diagnosis. The neoplasm was diagnosed as a WHO Grade IV Glioblastoma Multiforme. The tumor was noted as hypercellular with atypical looking cells. Microvascular proliferation, mitotic figures, and necrosis were also evident in the samples. The cells were positive for GFAP, confirming its glial origins. P53 staining was variable, with positivity seen in the majority of the tumor cells. The IDH1 R132H mutation and EGFR were both negative. Ki67 staining revealed between 30 to 40 percent of tumor cells were proliferating. The tumor was also positive for MGMT promoter methylation. The patient was lost to follow up and not a patient at the institution as of July 2014.
Discussion
Tissue Microarray Quality
Tissue microarrays are a very valuable tool. They enable researchers to examine many samples of tissue at once. By putting many samples on one slide, immunohistochemistry staining is made much more time and cost efficient. Up to 40 cases can be stained at once, maximizing the usage of the stain. However, not all cases can be used for a tissue microarray. I started with 208 cases, but only 82 made it to the two tissue microarrays. This is due to the fact that some tissue samples are simply insufficient for coring, being that they were too small or not optimal. I was unable to screen every glioblastoma case for the mutation. This is one of the disadvantages of using tissue microarrays to screen tumors for mutations.
The tissue microarrays I created were of good quality. 86.5% and 91% of the cores were either good or excellent and could be analyzed for the BRAF V600E mutation. Because tumor tissue samples are not uniform, slices of tissue in the array tend to vary from one cut to another. Although there were some cores that had insufficient tumor or were missing, almost all of the cases had at least one to two sufficient cores for analysis. The use of a tissue microarray to analyze the tissue samples was not only cost and time efficient, but it was a successful method.
Incidence of BRAF V600E Mutation in Tissue Microarray Glioblastoma Cohort
The BRAF V600E mutation occurs in a very low percentage of glioblastoma, although they appear in a higher percentage in certain subtypes and groups such as epithelioid glioblastoma and pediatric glioblastoma. In this study, I did not focus on a specific group of glioblastoma patients. I analyzed a cohort of all glioblastoma patients that were only selected based on the diagnosis of a WHO Grade IV astrocytic tumor.
I screened a total of 82 patients for the BRAF V600E mutation. I found four of the 82 to be positive by immunohistochemistry. This is 4.88% of the patients. However, sequencing confirmed that two of these four cases were positive for the mutation. Two of the 82 cases were truly positive for the BRAF V600E mutation. This is 2.45% of the patients. The stain was 50% accurate at revealing the mutation. The percent incidence of the BRAF V600E point mutation I found in my study was slightly lower than that of previous ones which found that the mutation can appear in up to 5% of glioblastoma.
The BRAF V600E Positive Cases
Finding the presence of the BRAF V600E mutation is very significant for the two patients. Glioblastoma is a very aggressive cancer and it is extremely likely that patients will have a recurrence at some point in their lives. Now that the mutation has been identified in these patients, they have a new treatment option available. Finding a new treatment option is very significant because glioblastoma patients are often out of treatment options. These patients may be treated with vemurafenib should they have a recurrence. This path of treatment has been successful in its application in few glioblastoma patients. It is promising that the new treatment option may prolong the survival of glioblastoma patients with few options.
Conclusion
Although the incidence of the BRAF mutation is relatively low in glioblastoma, the discovery of the mutation is significant because it can be targeted with an inhibitor drug that has been successful in its applications in glioblastoma patients. Glioblastoma patients are faced with a very aggressive cancer and a poor prognosis. There are few treatment options available and none are very promising. Adding to the list of possible treatments gives patients with the BRAF V600E mutation a better chance at prolonged survival. The two parents that I have diagnosed with the mutation now have another treatment option that they may pursue. This is very significant for the patients in particular, who may be out of options to combat their disease.
I have shown that tissue microarrays are an effective way of identifying the BRAF V600E mutation in glioblastoma. Staining individual slides of cases and/or sequencing every case that needs to be screened is not only time consuming, but costly. The BRAF V600E mutation occurs at a low incidence rate in glioblastoma, so screening each case by immunohistochemistry or sequencing individually is not ideal or efficient. Tissue microarrays allow a stain to be maximized and many tumors to be screened at once. The arrays I compiled were of excellent quality and most of the cores were intact and able to be stained. I may now say that all glioblastoma cases should be screened for the BRAF mutation by immunohistochemistry staining of tissue microarrays. The cases that are positive can be verified with sequencing and patients may be given a new treatment option that may save their life. Although the rate of occurrence of the BRAF V600E mutation in glioblastoma is relatively low, patients should be screened because a successful targeted therapy is available. In addition, the tissue microarrays used to screen for BRAF may be used to screen for other mutations in the same way.
There is still a lot that is unknown about the BRAF V600E mutation in the context of glioblastoma multiforme. There are no known histological or clinical features that differentiate BRAF V600E positive and negative cases. This would be a further focus of mine. In the future, I aim to identify a characteristic that may indicate the presence of a BRAF V600E point mutation in a tumor sample. I would gather a much larger cohort of glioblastoma that are known to have the mutation for this study and look at histological features, other mutations or gene amplifications, and the demographic characteristics of the patients themselves.
References
Brandner, S., and A. Von Deimling. "Diagnostic, Prognostic, and Predictive Relevance of Molecular Markers in Gliomas." Neuropathology and Applied Neurobiology (2015): 1-25. PubMed. Web. 19 Aug. 2015.
Dias-Santagata, Dora, and Et al., "BRAF V600E Mutations Are Common in Pleomorphic Xanthoastrocytoma: Diagnostic and Therapeutic Implications." PLoS One 6.3 (2011): 1-9. PubMed. Web. 19 Aug. 2015.
"Glioblastoma Multiforme." American Association of Neurological Surgeons. N.p., n.d. Web. 8 Nov. 2015. <http://www.aans.org/patient%20information/conditions%20and%20treatments/glioblastoma%20multiforme.aspx>.
Gutmann, David H. "BRAF V600E Mutation in Pediatric and Adult Glioblastoma." Neuro Oncology (2013): 318-19. PubMed. Web. 19 Aug. 2015.
Kanitakis, Jean. "Novel Approaches to Treatment of Advanced Melanoma: A Review on Targeted Therapy and Immunotherapy." BioMed Research International (2015): n. pag. PubMed. Web. 27 Aug. 2015.
Kleinschmidt De-Masters, Bette K. "Epithelioid Glioblastomas Show a High Percentage of BRAF V600E Mutation." The American Journal of Surgical Pathology 37 (2013): 685-97. PubMed. Web. 19 Aug. 2015.
Kleinschmidt-DeMasters, Bette K., and Et al., "BRAF VE1 Immunoreactivity Patterns in Epithelioid Glioblastomas Positive for BRAF V600E Mutation." American Journal of Surgical Neuropathology 39.4 (2015): 528-40. PubMed. Web. 19 Aug. 2015.
Love, Seth, and David Ellison. "Glioblastoma." A Reference Text of CNS Pathology. By Love and
Ellison. N.p.: Elsevier Limited, 2013. N. pag. Print.
Nicolaides, Theodore P., and Et al., "Targeted Therapy for BRAF V600E Malignant Astrocytoma." National Institute of Health (2011): 1-21. PubMed. Web. 19 Aug. 2015.
Prados, Michael D., and Et al., "Toward Precision Medicine in Glioblastoma: The Promise and The Challenges." Neuro-Oncology (2015): 1051-63. PubMed. Web. 19 Aug. 2015.
Robinson, Giles W. "Complete Clinical Regression of a BRAF V600E Mutant Pediatric Glioblastoma Multiforme After BRAF Inhibitor Therapy." BMC Cancer 14.258 (2014): 1-5. PubMed. Web. 27 Aug. 2015.
Schindler, Genevieve, and Et al., "Analysis of BRAF V600E Mutation in 1,320 Nervous System Tumors Reveals High Mutation Frequencies in Pleomorphic Xanthoastrocytoma, Ganglioglioma and Extra-cerebellar pilocytic astrocytoma." Springer-Verlag (2011): 397-405. PubMed. Web. 19 Aug. 2015.


Post a comment